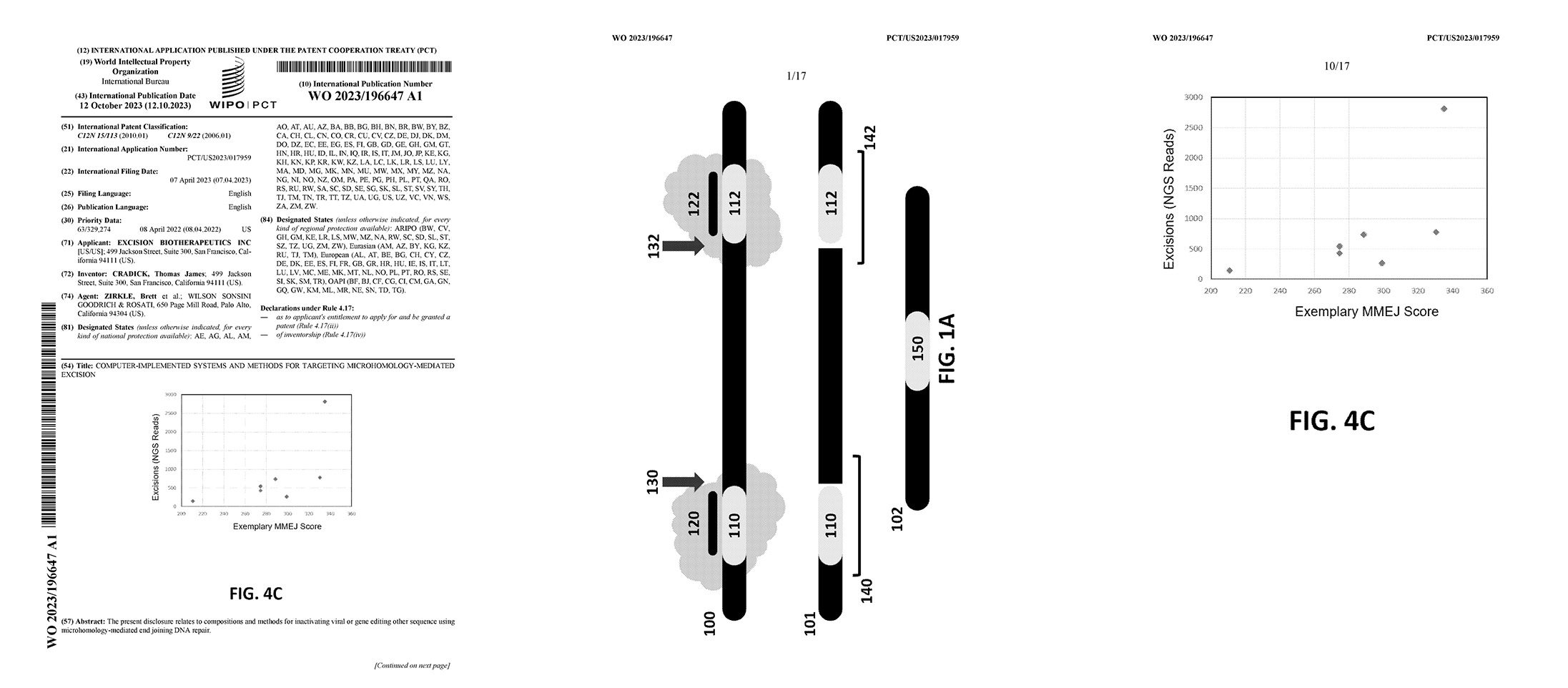
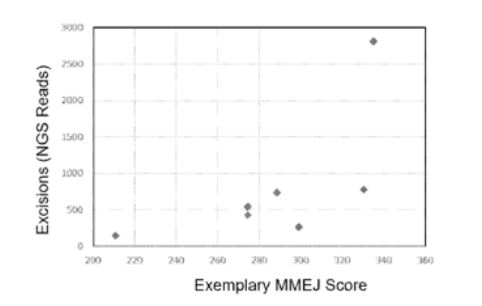
Compositions and methods for inactivating viral targets or gene editing other sequences using microhomology-mediated end joining (MMEJ) DNA repair. For example, the described methods better allow for the selection of targets sites predicated to provide a higher rate of the desired outcome.
S Police, Chou, TJ Cradick, R Ng, & Y Yang
Cas systems and compositions that target the dystrophin gene. Also provided are methods for using the CRISPR/Cas systems, vectors and compositions in methods for genome engineering to correct a mutant dystrophin gene, and for treating Duchenne muscular dystrophy.
Seshi Police, Song Chou, TJ Cradick, & Robert Ng
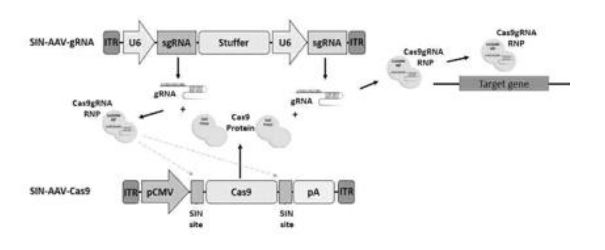
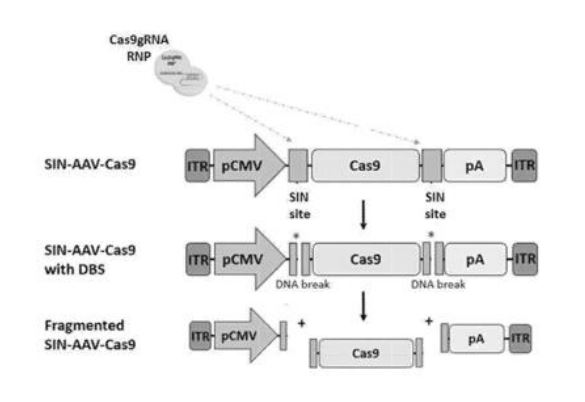
The invention relates to self-inactivating/self-targeting CRISPR-Cas or CRISPR-CPFL systems, to related genetically modified cells, and to methods of controlling Cas9 expression. The invention also relates to methods of genetically modifying a cell, to nucleic acids for use in a self-inactivating CRISPR/Cas or CRISPR/Cpfl system and to related pharmaceutical compositions.
TJ Cradick, C Cowan, AS Lundberg, & G Cost
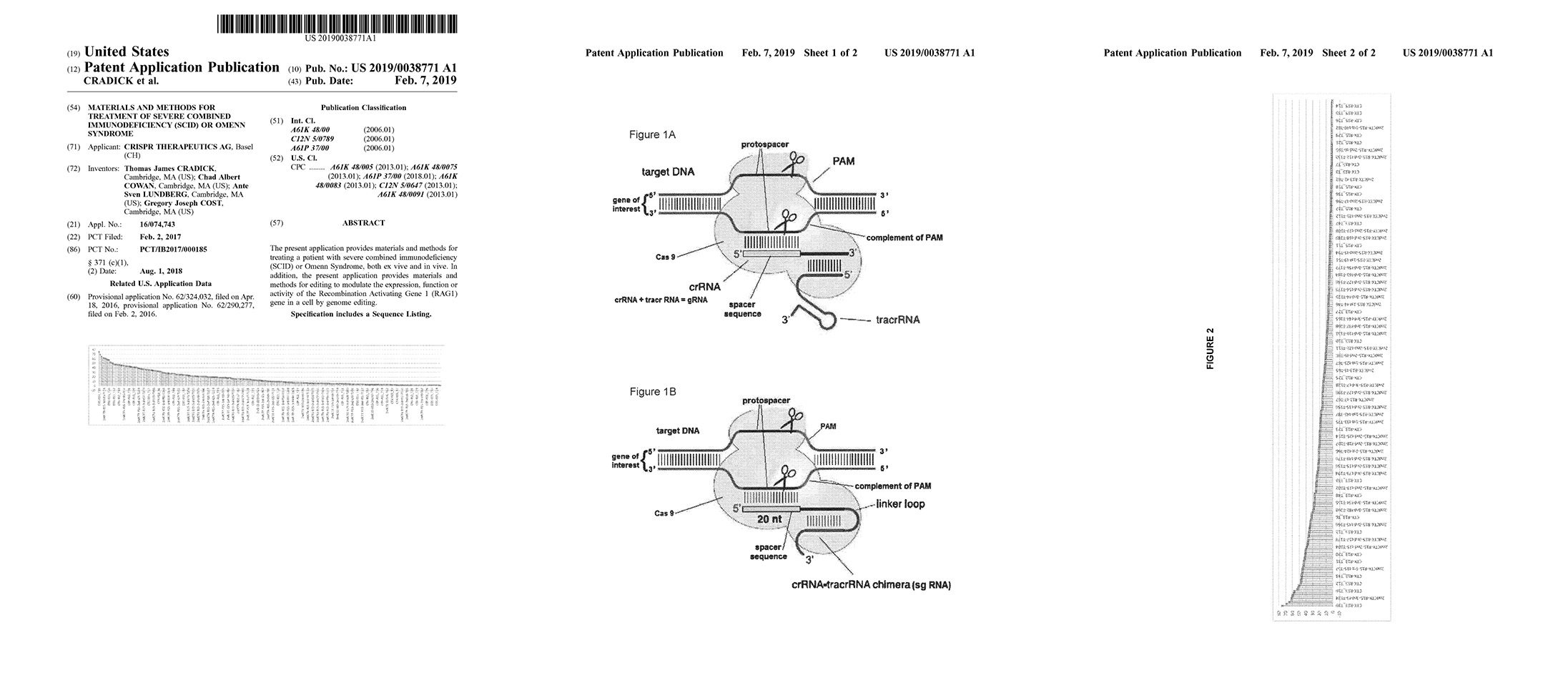
Materials and methods for treating a patient with severe combined immunodeficiency (SCID) or Omenn Syndrome, both ex vivo and in vivo. In addition, the present application provides materials and methods for editing to modulate the expression, function or activity of the Recombination Activating Gene 1 (RAG1) gene by genome editing.
Matt Porteus, TJ Cradick, G Bao, & C Lee
Materials and methods for treating hemoglobinopathies. More specifically, the application provides methods for producing progenitor cells that are genetically modified via genome editing to increase the production of fetal hemoglobin (HbF), as well as modified progenitor cells (including, for example, CD34+ human hematopoietic stem cells) producing increased levels of HbF, and methods of using such cells for treating hemoglobinopathies such as sickle cell anemia and B-thalassemia.
TJ Cradick, P Qiu, & G Bao
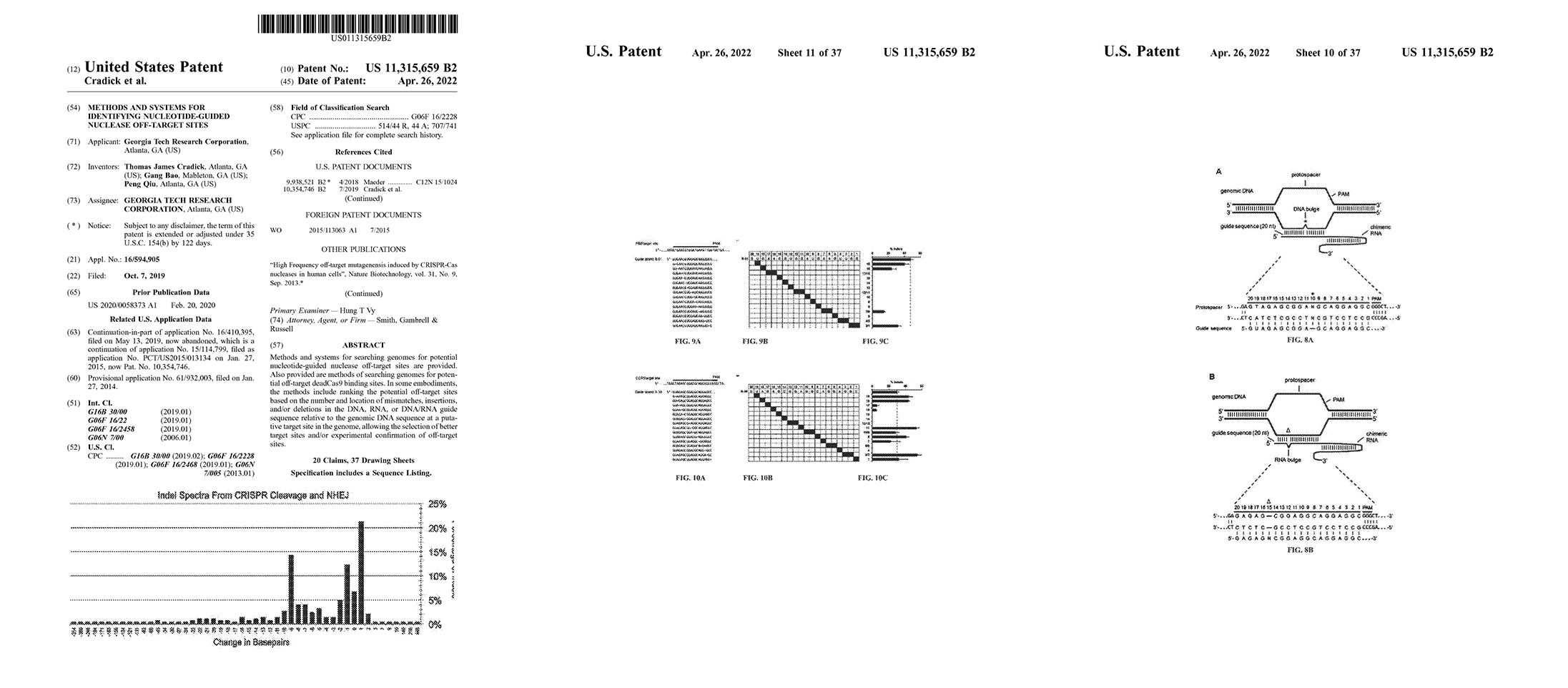
Licensed patent covering methods and systems for searching genomes for potential CRISPR off-target sites based on the number and location of mismatches, insertions, and/or deletions in the gRNA guide sequence relative to the genomic DNA sequence at a putative target site in the genome. (This covers searching for sites with bulges or gaps between the guide RNA and DNA sequences).
These methods allow for the selection of better target sites and/or experimental confirmation of off-target sites and are an improvement over partial search mechanisms that fail to locate every possible target site.
EJ Fine, TJ Cradick, Y Lin, & G Bao.
Provided herein are systems and methods for identifying the off-site cleavage loci and predicting the activity of engineered endonucleases for a given genome. It is expected that these tools and methods will be useful for designing nucleases and other related DNA binding domains (e.g. TAL effectors) for genomic therapy and engineering.
AP McCaffrey & TJ Cradick
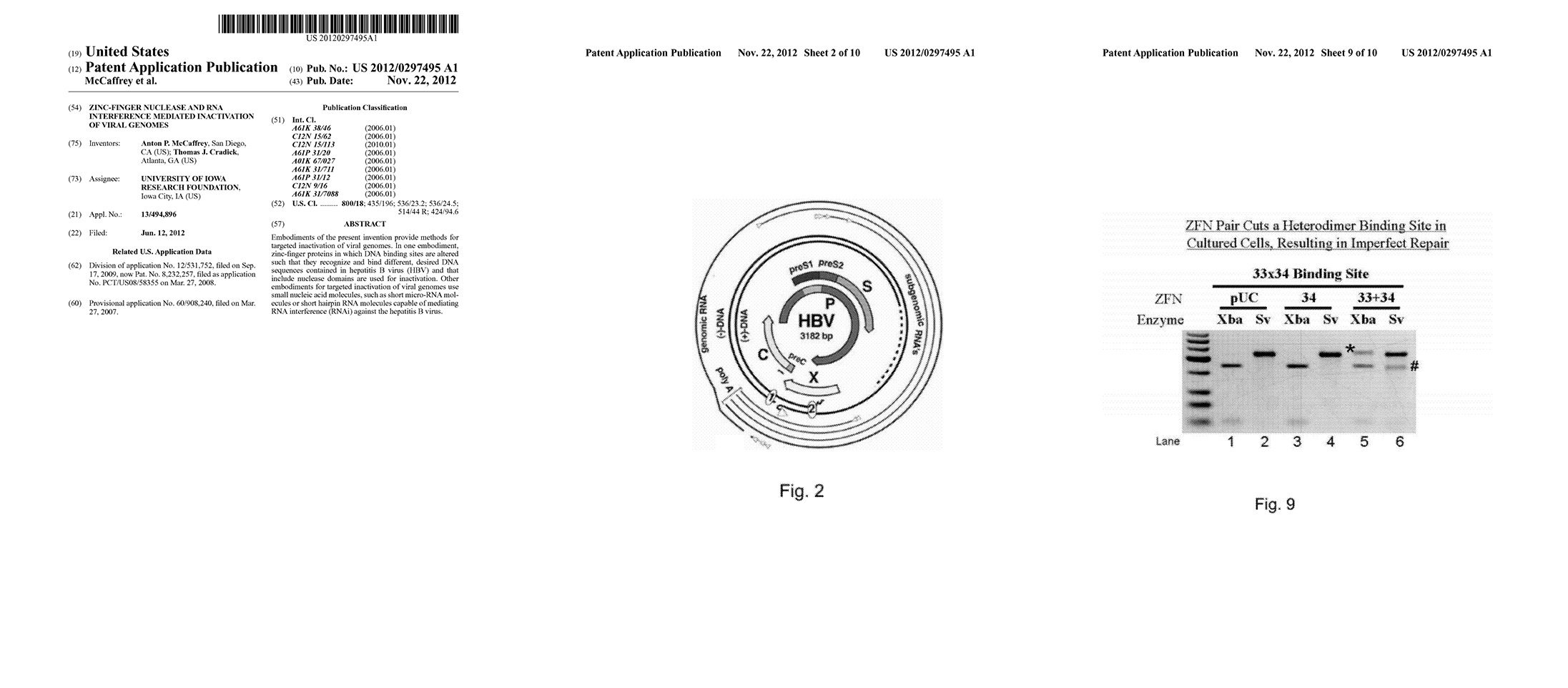
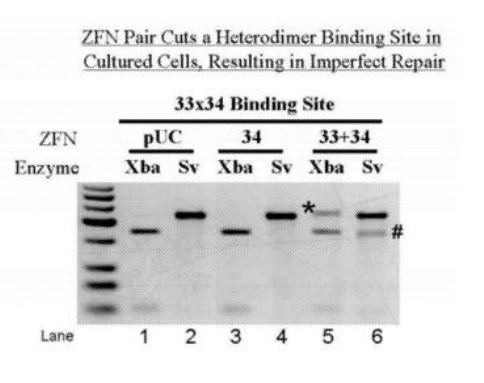
ZFN Pair Cuts a Heterodimer Binding Site in Cultured Cells, Resulting in Imperfect Repair
Methods for targeted inactivation of viral genomes. In one embodiment, zinc-finger proteins in which DNA binding sites are altered such that they recognize and bind different, desired DNA sequences contained in hepatitis B virus (HBV) and that include nuclease domains are used for inactivation (Zinc Finger Nucleases = ZFNs). Other embodiments for targeted inactivation of viral genomes use small nucleic acid molecules, such as short micro-RNA molecules or short hairpin RNA molecules capable of mediating RNA interference (RNAi) against HBV.
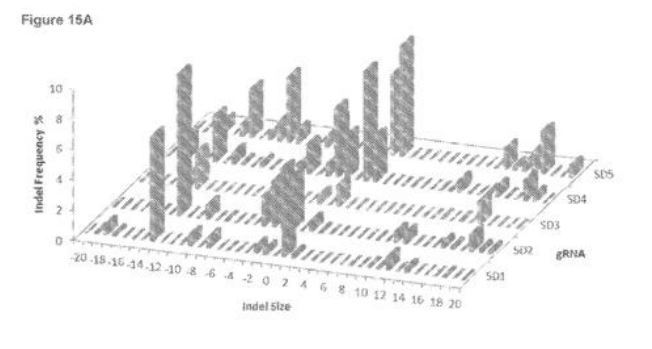
2024 Computer-Implemented Systems & Methods for Targeting Microhomology-Mediated Excision
WIPO (PCT) WO2023196647A1
TJ Cradick
Compositions and methods for inactivating viral targets or gene editing other sequences using microhomology-mediated end joining (MMEJ) DNA repair. For example, the described methods better allow for the selection of targets sites predicated to provide a higher rate of the desired outcome.
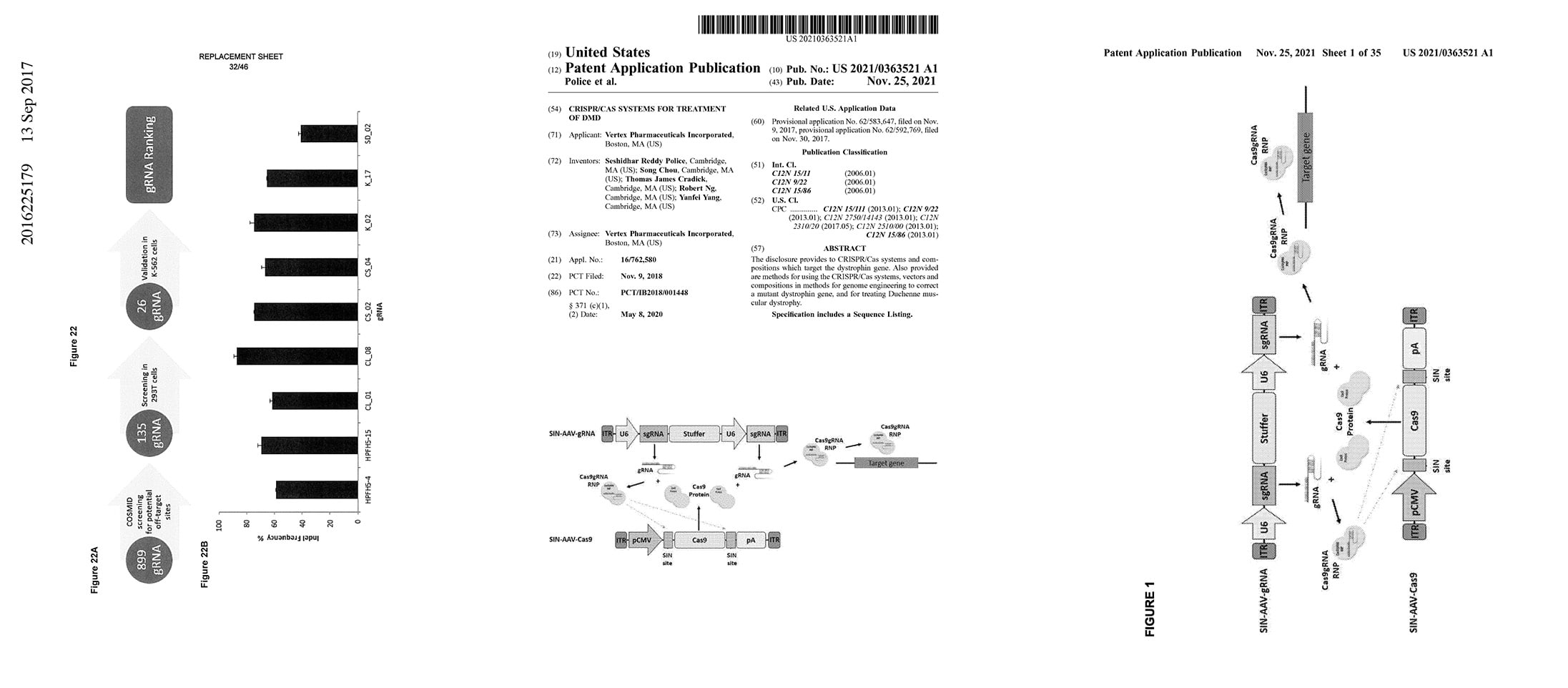
2021 CRISPR/CAS Systems for Treatment of DMD
US20210363521A1
S Police, Chou, TJ Cradick, R Ng, & Y Yang
Cas systems and compositions that target the dystrophin gene. Also provided are methods for using the CRISPR/Cas systems, vectors and compositions in methods for genome engineering to correct a mutant dystrophin gene, and for treating Duchenne muscular dystrophy.
2019 Self-Inactivating (SIN) CRISPR/Cas or CRISPR/Cpf1 Systems & Uses Thereof
Patent Application WO2019092505A1
Seshi Police, Song Chou, TJ Cradick, & Robert Ng
The invention relates to self-inactivating/self-targeting CRISPR-Cas or CRISPR-CPFL systems, to related genetically modified cells, and to methods of controlling Cas9 expression. The invention also relates to methods of genetically modifying a cell, to nucleic acids for use in a self-inactivating CRISPR/Cas or CRISPR/Cpfl system and to related pharmaceutical compositions.
2017 Materials & Methods for Treatment of Severe Combined Immunodeficiency (SCID) or Omenn Syndrome
TJ Cradick, C Cowan, AS Lundberg, & G Cost
Materials and methods for treating a patient with severe combined immunodeficiency (SCID) or Omenn Syndrome, both ex vivo and in vivo. In addition, the present application provides materials and methods for editing to modulate the expression, function or activity of the Recombination Activating Gene 1 (RAG1) gene by genome editing.
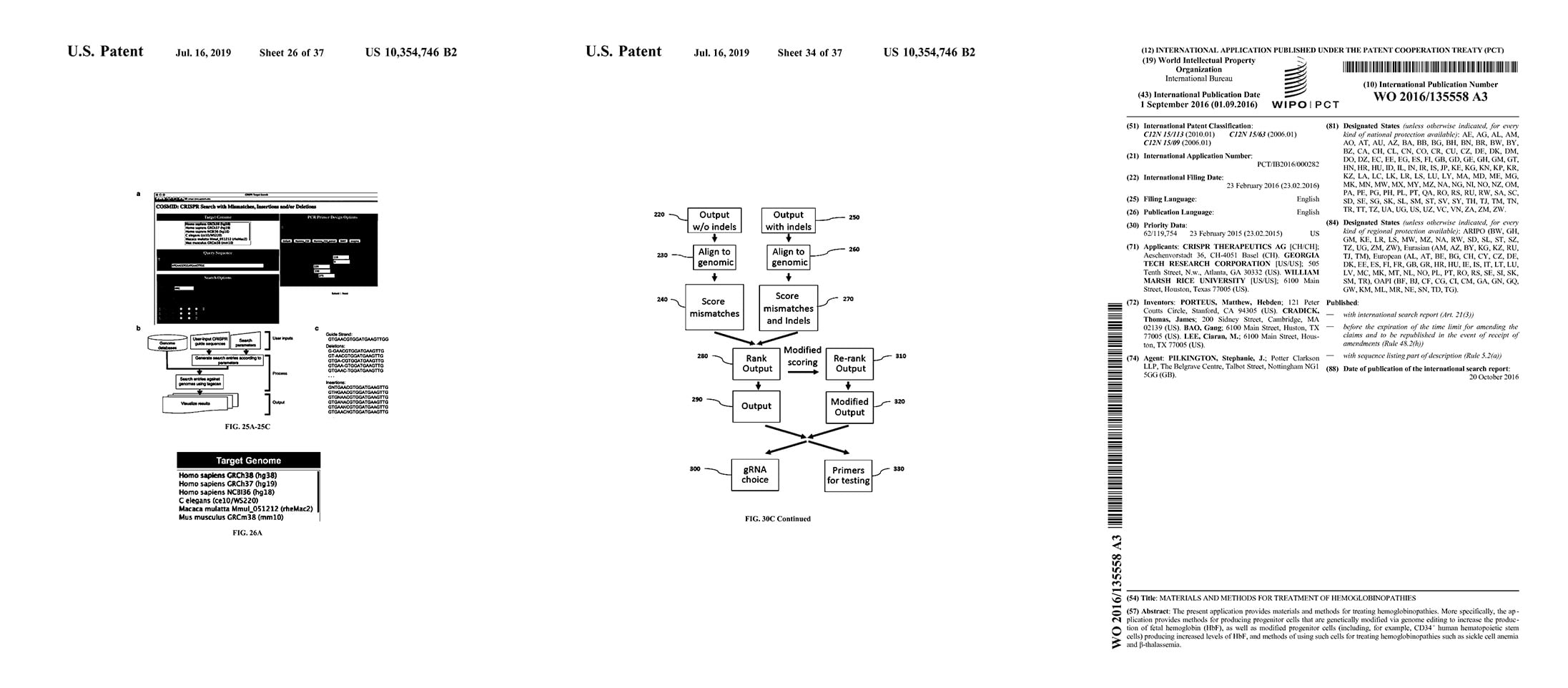
2016 Materials & Methods for Treatment of Hemoglobinopathies
Matt Porteus, TJ Cradick, G Bao, & C Lee
Materials and methods for treating hemoglobinopathies. More specifically, the application provides methods for producing progenitor cells that are genetically modified via genome editing to increase the production of fetal hemoglobin (HbF), as well as modified progenitor cells (including, for example, CD34+ human hematopoietic stem cells) producing increased levels of HbF, and methods of using such cells for treating hemoglobinopathies such as sickle cell anemia and B-thalassemia.
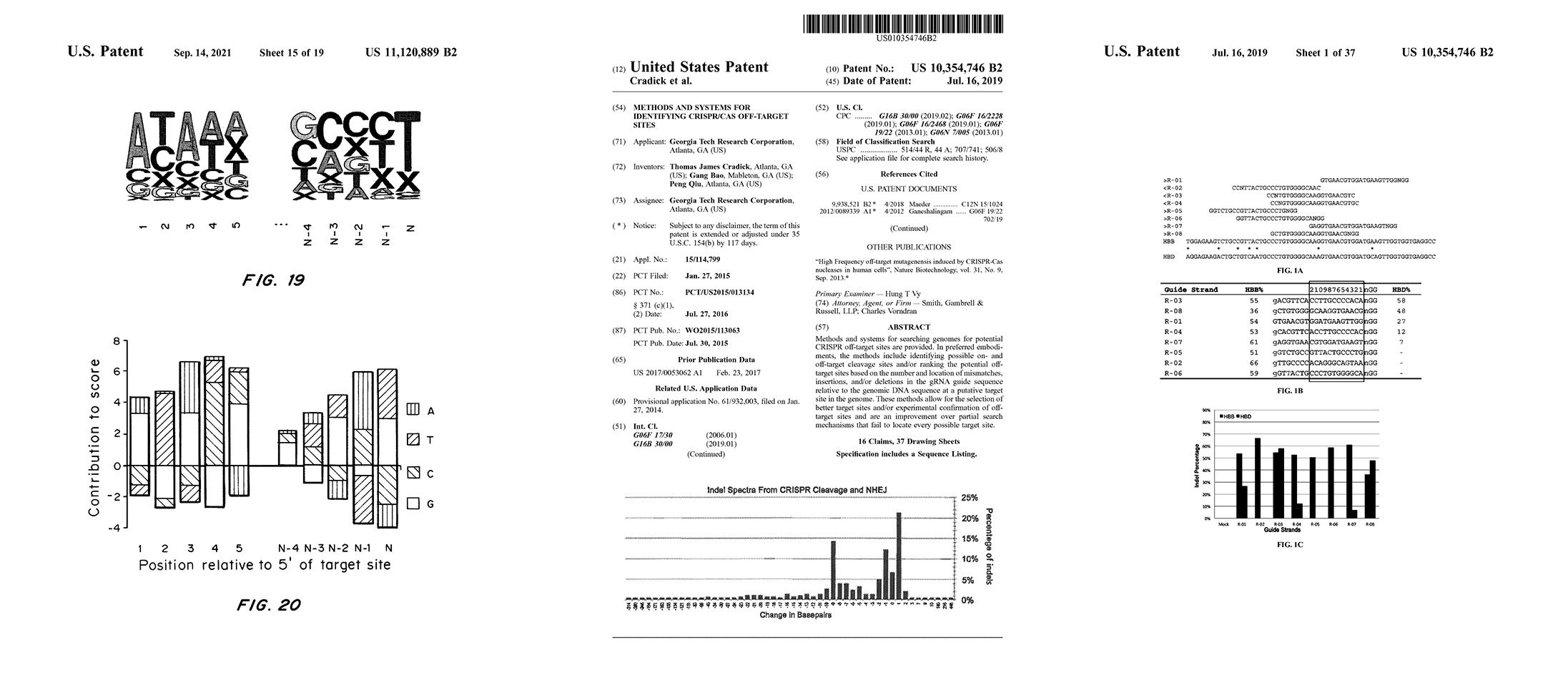
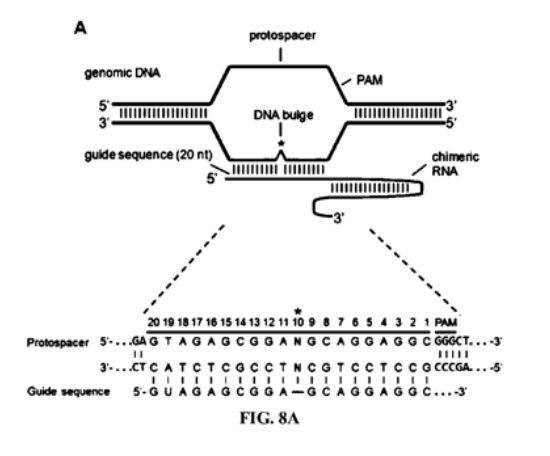
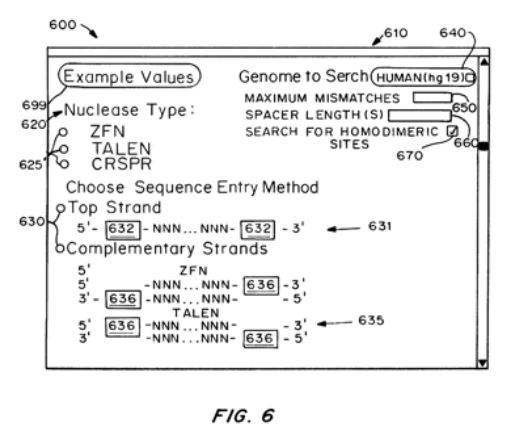
2014 Methods & Systems for Identifying CRISPR/Cas Off-Target Sites
US10354746B2
WO2015113063A1
US11315659B2
TJ Cradick, P Qiu, & G Bao
Licensed patent covering methods and systems for searching genomes for potential CRISPR off-target sites based on the number and location of mismatches, insertions, and/or deletions in the gRNA guide sequence relative to the genomic DNA sequence at a putative target site in the genome. (This covers searching for sites with bulges or gaps between the guide RNA and DNA sequences).
These methods allow for the selection of better target sites and/or experimental confirmation of off-target sites and are an improvement over partial search mechanisms that fail to locate every possible target site.
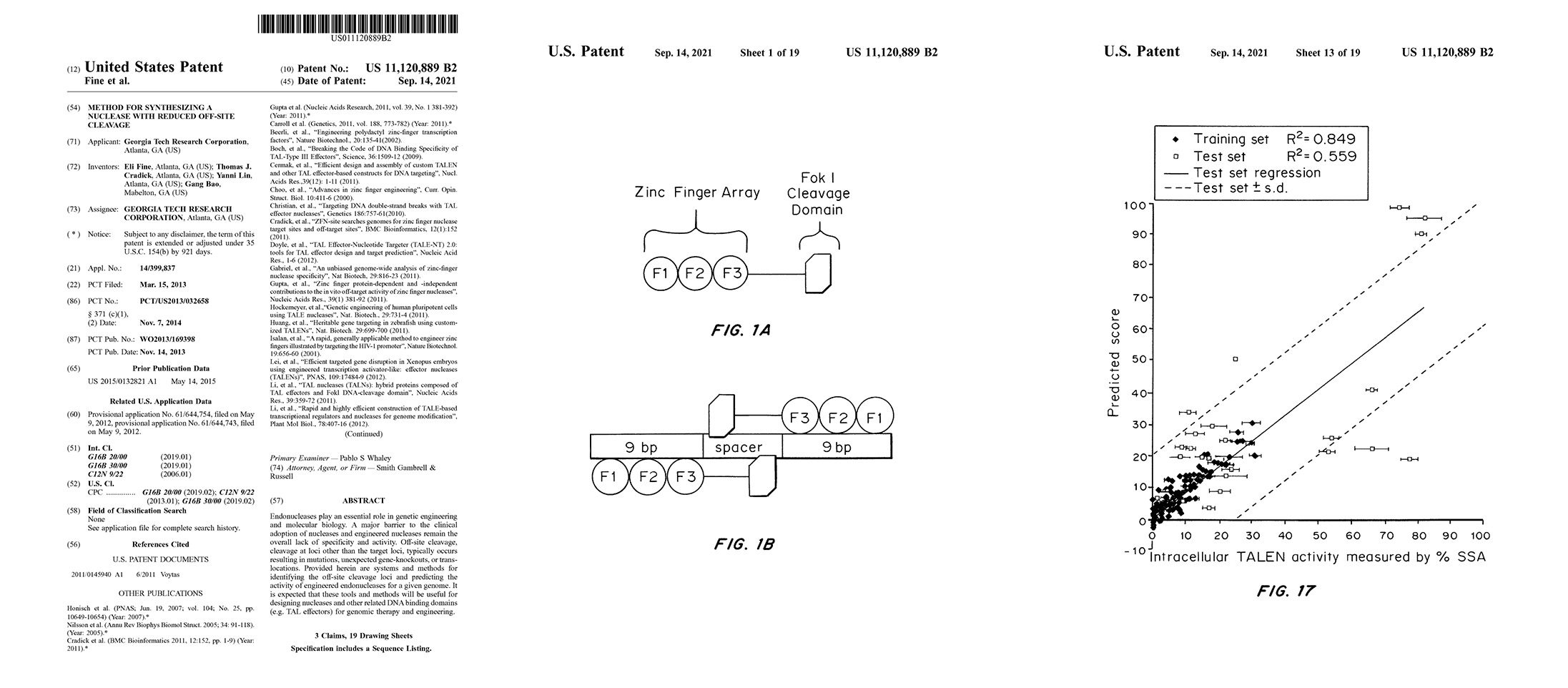
2013 Method for Synthesizing a Nuclease with Reduced Off-Site Cleavage
US11120889B2
EJ Fine, TJ Cradick, Y Lin, & G Bao.
Provided herein are systems and methods for identifying the off-site cleavage loci and predicting the activity of engineered endonucleases for a given genome. It is expected that these tools and methods will be useful for designing nucleases and other related DNA binding domains (e.g. TAL effectors) for genomic therapy and engineering.
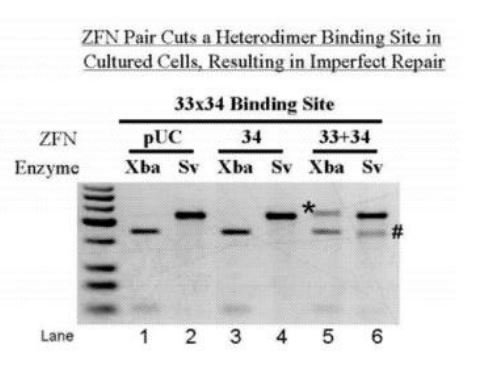
2012 Zinc-Finger Nuclease & RNA Interference-Mediated Inactivation of Viral Genomes
US20120297495A1
AP McCaffrey & TJ Cradick
ZFN Pair Cuts a Heterodimer Binding Site in Cultured Cells, Resulting in Imperfect Repair
Methods for targeted inactivation of viral genomes. In one embodiment, zinc-finger proteins in which DNA binding sites are altered such that they recognize and bind different, desired DNA sequences contained in hepatitis B virus (HBV) and that include nuclease domains are used for inactivation (Zinc Finger Nucleases = ZFNs). Other embodiments for targeted inactivation of viral genomes use small nucleic acid molecules, such as short micro-RNA molecules or short hairpin RNA molecules capable of mediating RNA interference (RNAi) against HBV.